Our solutions are aimed at reducing or eliminating the prevalence and load of microorganisms related to infections and diseases in intensive animal farming.
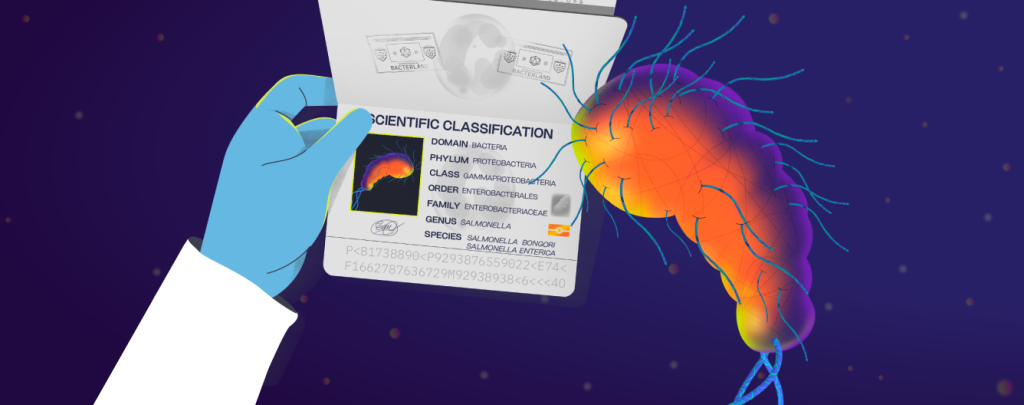
We obtain strains of various serovars from breeding and rearing sectors, feed mills and slaughter plants. These are characterized at genomic level to obtain detailed information on their virulence profiles, antibiotic resistance, and their distribution in different stages and sectors of production.
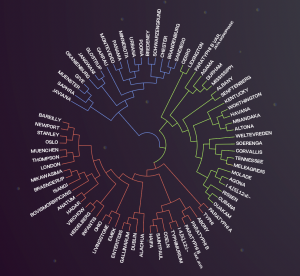
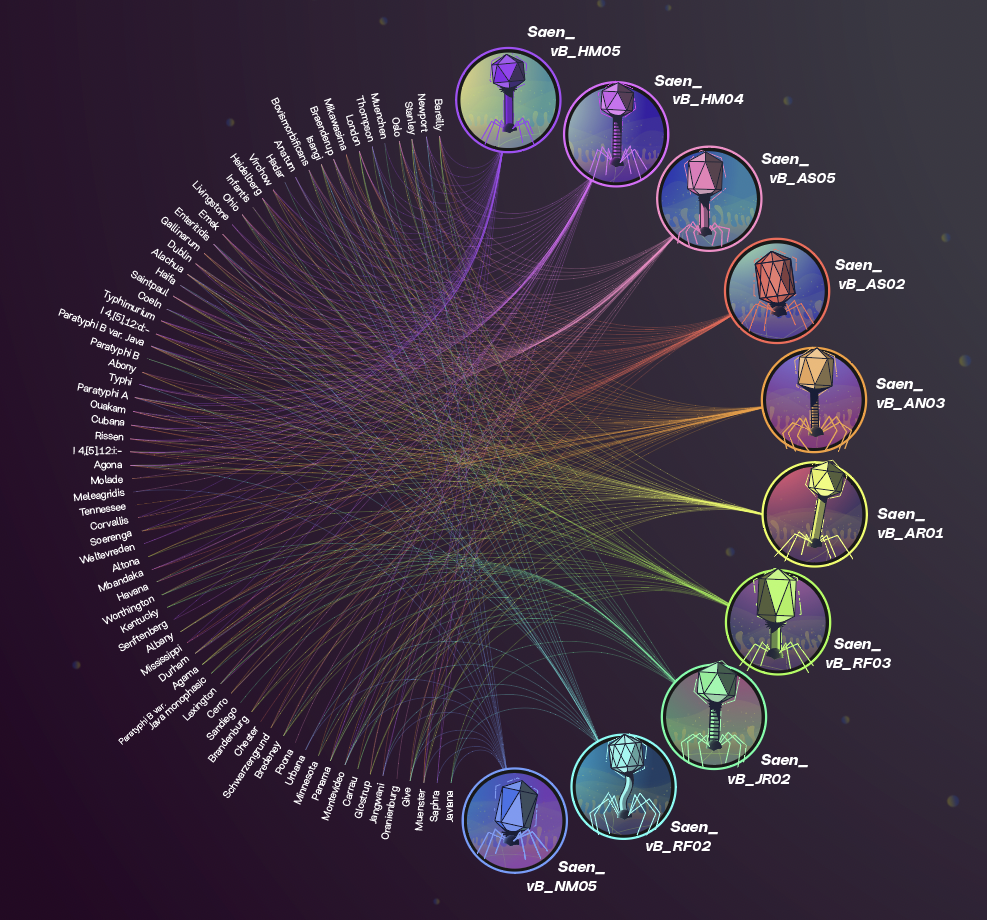
The simultaneous comparison of all genes in all isolates allows us to determine phylogenetic relationships between them, in terms of proximity and remoteness, being able to infer relationship exchanges in species or bacterial groups between the different sectors and productive stages.
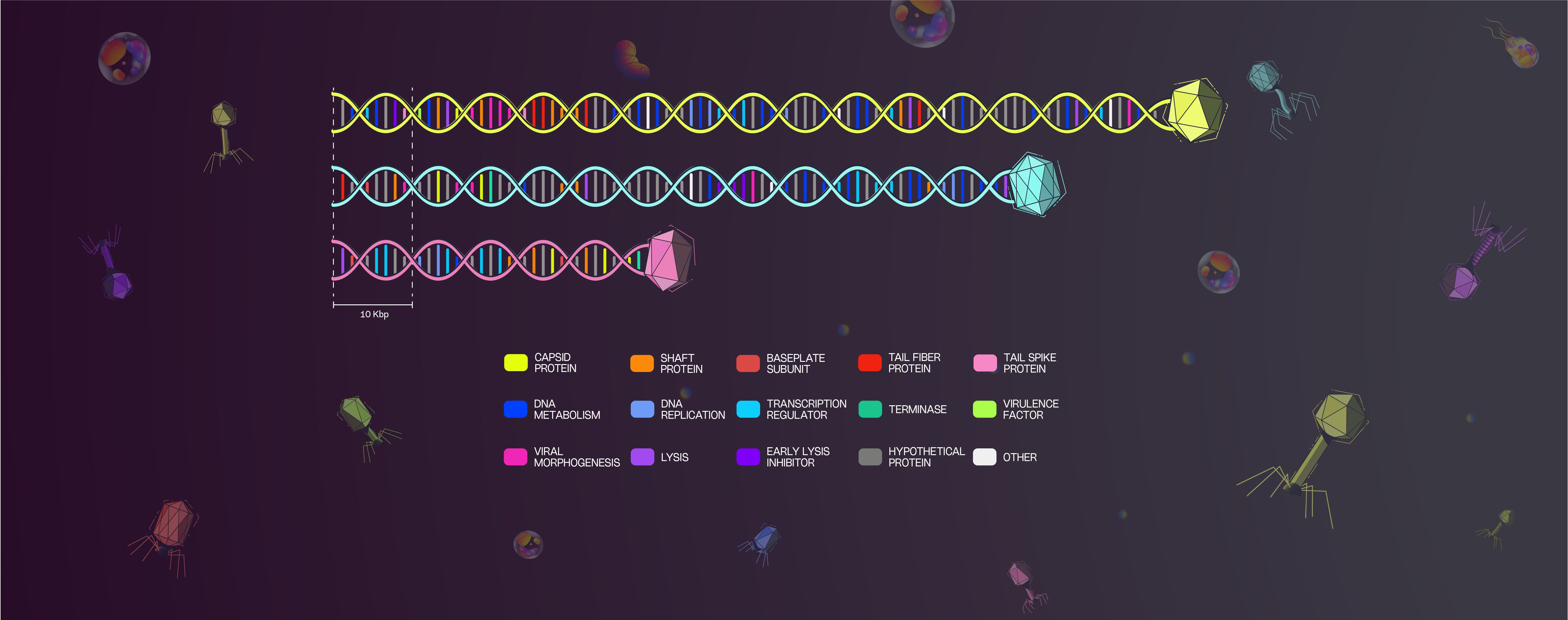
Each predicted gene was assigned to one of twenty categories, which are composed of proteins that perform similar biological functions. Subsequently, we generate a map illustrating the spatial organization of functional genes within the genome of each bacteriophage. All of our bacteriophages have a lytic replicative cycle. Also, we guarantee the absence of elements related to a potential propagation of bacterial genetic material (such as integrases, recombination sites, bacterial origin, virulence and resistance genes).
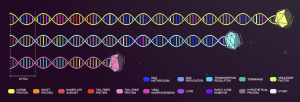
All these characteristics imply that the candidate bacteriophages are suitable for use in phage therapy, in accordance to the biosafety of humans, animals and the environment.
The antimicrobial effect of bacteriophages on the isolates is determined by comparing the relationship between the area under the growth curve in the presence and absence of the bacteriophage. Null effect will be inhibition up to 15% of growth. Partial inhibition will be between 15% and 85%, and total inhibition will be a reduction over 85%.
Thousands of tons of chicken meat test positive for Salmonella even after cooking, posing a danger to human population.
We propose the use of bacteriophage therapy to reduce the load and prevalence of Salmonella throughout the production chain, both in breeding and slaughter.